【*】AI Go Senior 补充 (2021)
环境安装
在使用Python开发AI时,由於需时时查看处理中的训练资料,於是大多使用Jupyter Notebook进行开发。这是利用Python是直译式程序语言的的特点,在这里就不说太多,到底要如何安装Jupyter Notebook呢?
Google Colab
优点:免下载、已预先安装大多数的函式库、搞不好速度比你家电脑快、档案会自动清除。
缺点:若想储存资料须将资料下载或将Colab挂接到Google Drive、说不定比你家电脑慢、不能长时间闲置(会停止运算,但档案还在)
使用
点开连结:Colab
首次登入介面:
(未登入Google)
嘿对~我Windows还没启用,但我其实有买,这就是另一个故事了~
(已登入Google)
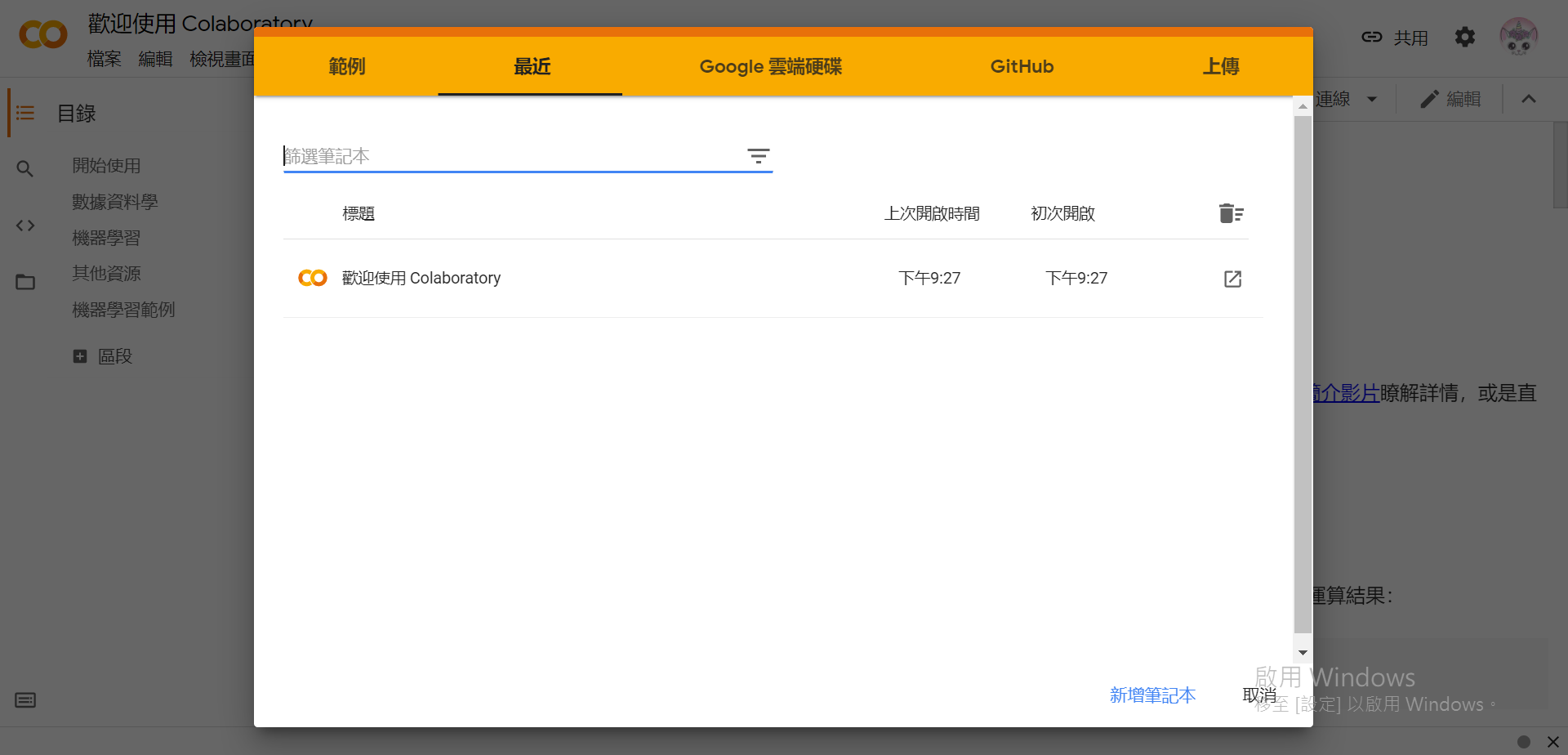
一共有5+1个选项:
- 范例:打开预设的范例程序
- 最近:可以快速存取最近的程序
- Google 云端硬碟:在Google Drive内打开(会自动存档)
- GitHub:复制GitHub上的专案
- 上传:上传并打开本地的档案(通常Jupyter Notebook会以
.ipynb作为副档名)。 - 取消:在右下角,直接跳过这个步骤并使用预设的程序范例。(不会自动存档,视窗关掉就没了)
这里我们选择[取消],如果要储存档案的话可以之後在左上方的[档案]中选择储存的位置。
如果觉得预设的范例很丑,可以点选第一个区块(我们称之为Cell),然後按垃圾桶进行删除。
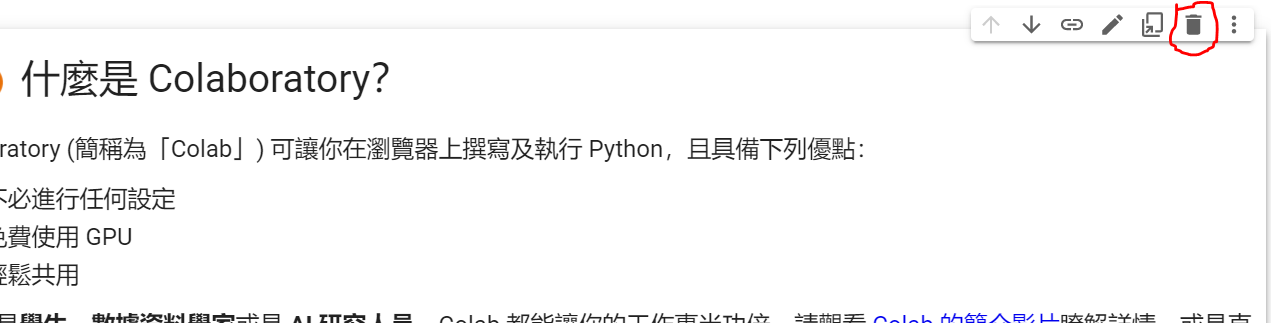
经一阵猛点後:
接着可以新增一个[+程序码](Python)或[文字](MarkDown)
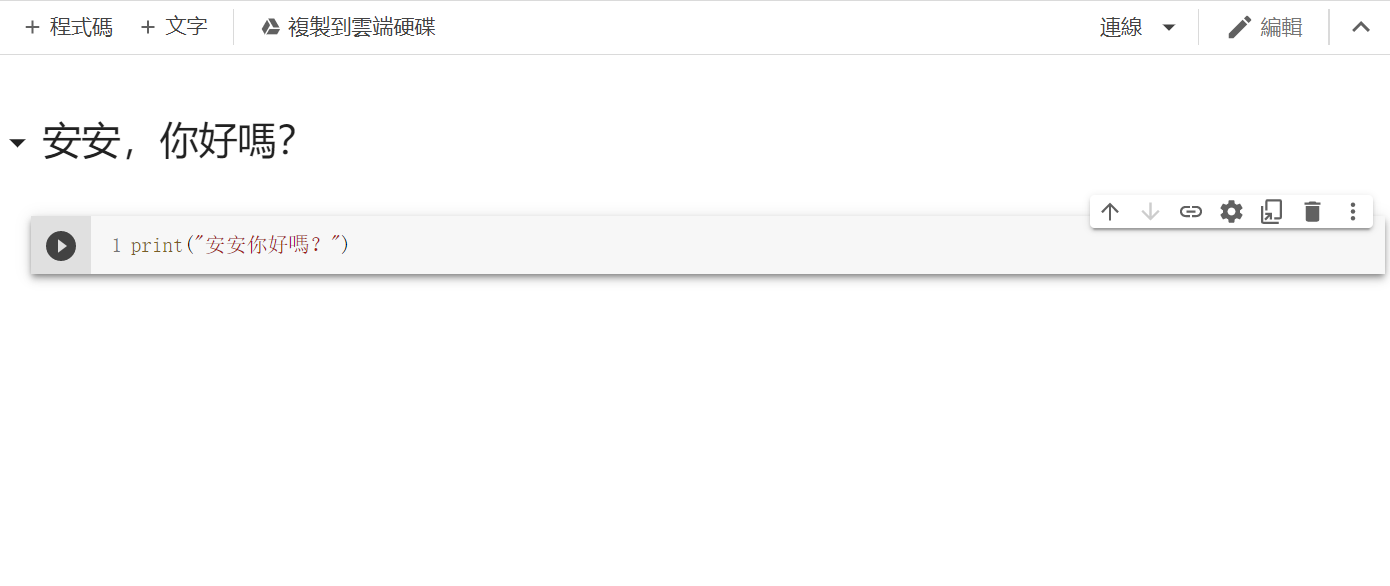
P.S:可以透过Cell右上方的箭头来调整Cell的顺序、
MarkDown的话可以点击[关闭Markdown编辑器](在设定旁边)来显示内容。
再来点及右上方共用底下有一个连线,点他来连线到主机後,点选程序码区块左边的执行储存格。
结果:
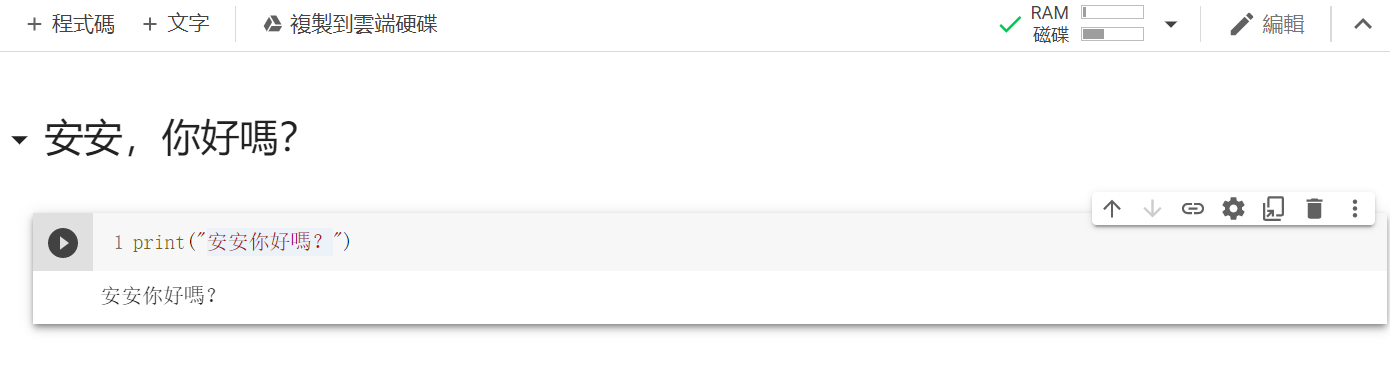
Google的Colab先到这边,剩下的未来遇到再说,也可以先自行探索喔~(例如使用GPU加速、Google特有的TPU等等)
Anaconda
如果不想执行在云端的话,另一个我推荐的是使用Anaconda IDE,他整合了许多资料科学套件,除了跑得有点慢,他会是你资料科学之旅的好朋友。
安装与建立环境
下载连结:Anaconda
能选64位元就选64位元,不然大量的数据灌下去你的电脑不知道要算到牛年马月。(当然这取决於作业系统及硬体配置,详细方法请见:如何查询你的Windows系统是32或64位元?)
安装完成後(就狂点下一步),初始画面如下:

接着点选Environments
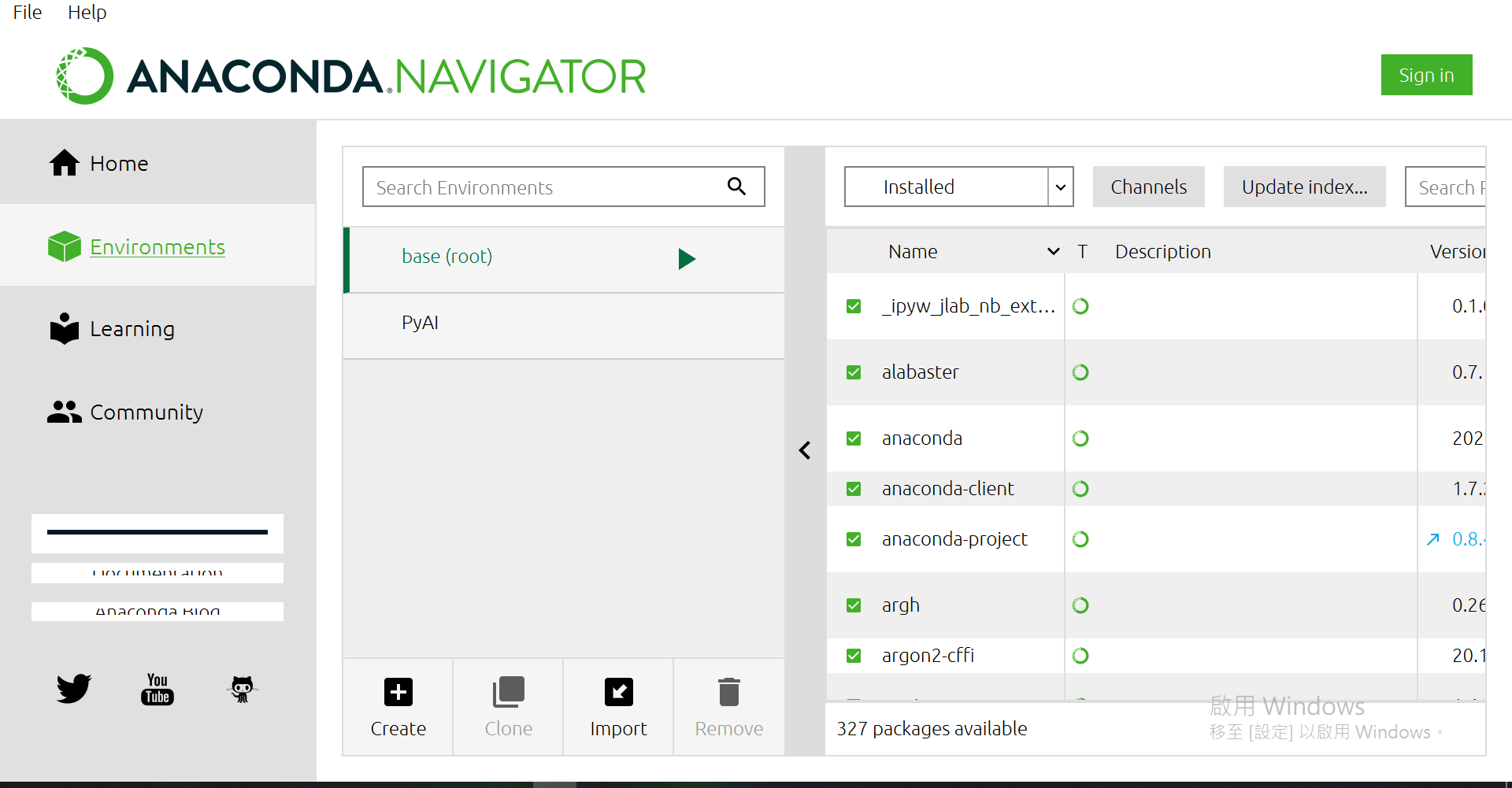
当然,你的画面会没有"PyAI"这个虚拟环境,现在就来创一个吧~
点选[Create]
打上自己想要的名子,版本我是选3.8啦~
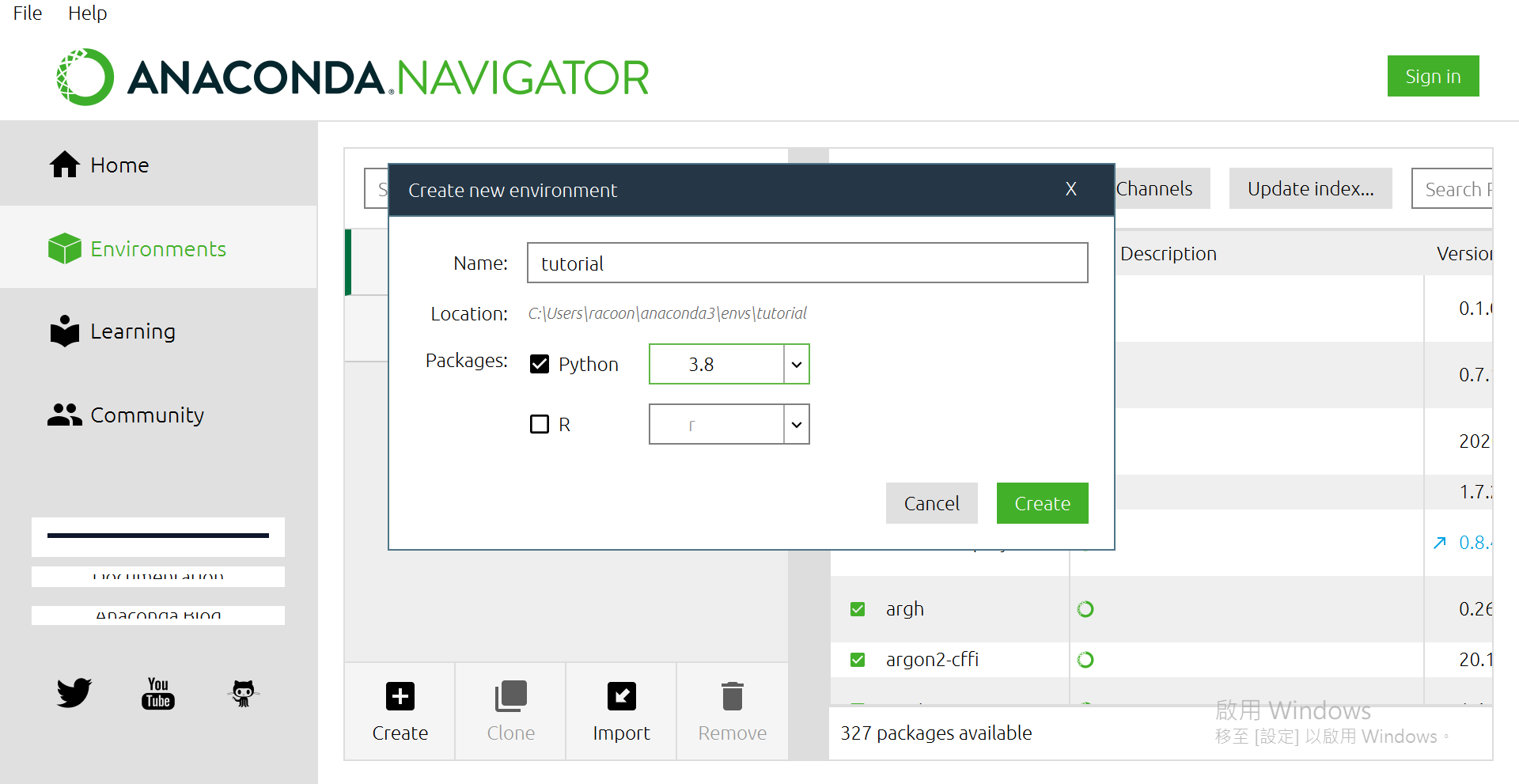
然後按[Create]
之後要选环境就可以进来这里选,或是在首页的地方有个"Applications on"可以切换。
接着安装未来会用到的函式库,保持在"Environments",中间有个箭头"<"给他按下去。
有一个选单"Installed"选成"Not Installed",再来右边可以搜寻,找到下面这几个package给他点选起来:
- scikit-learn
- pandas
- keras
- matplotlib
- request
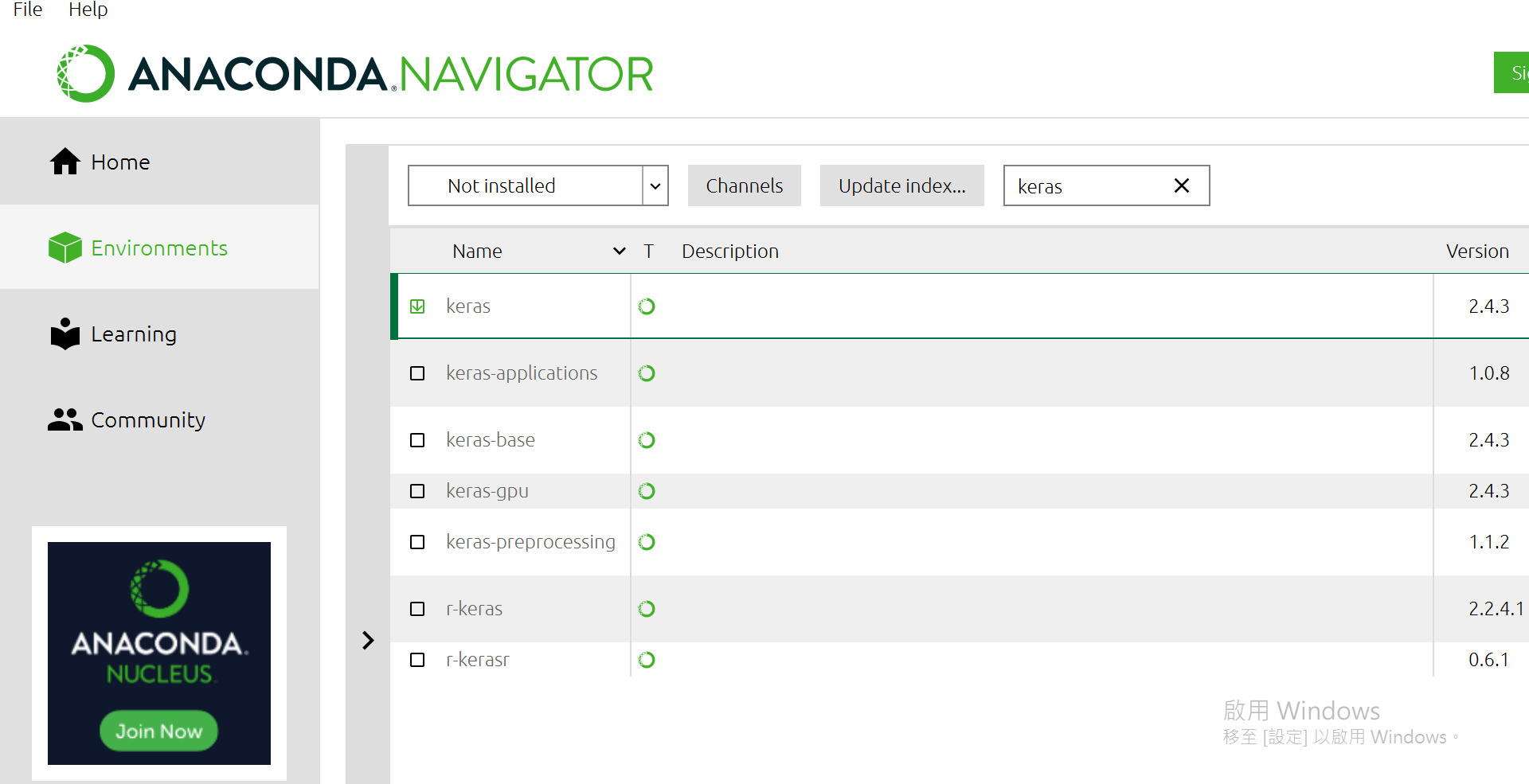
都选完後,右下方有个[Apply],按下去。
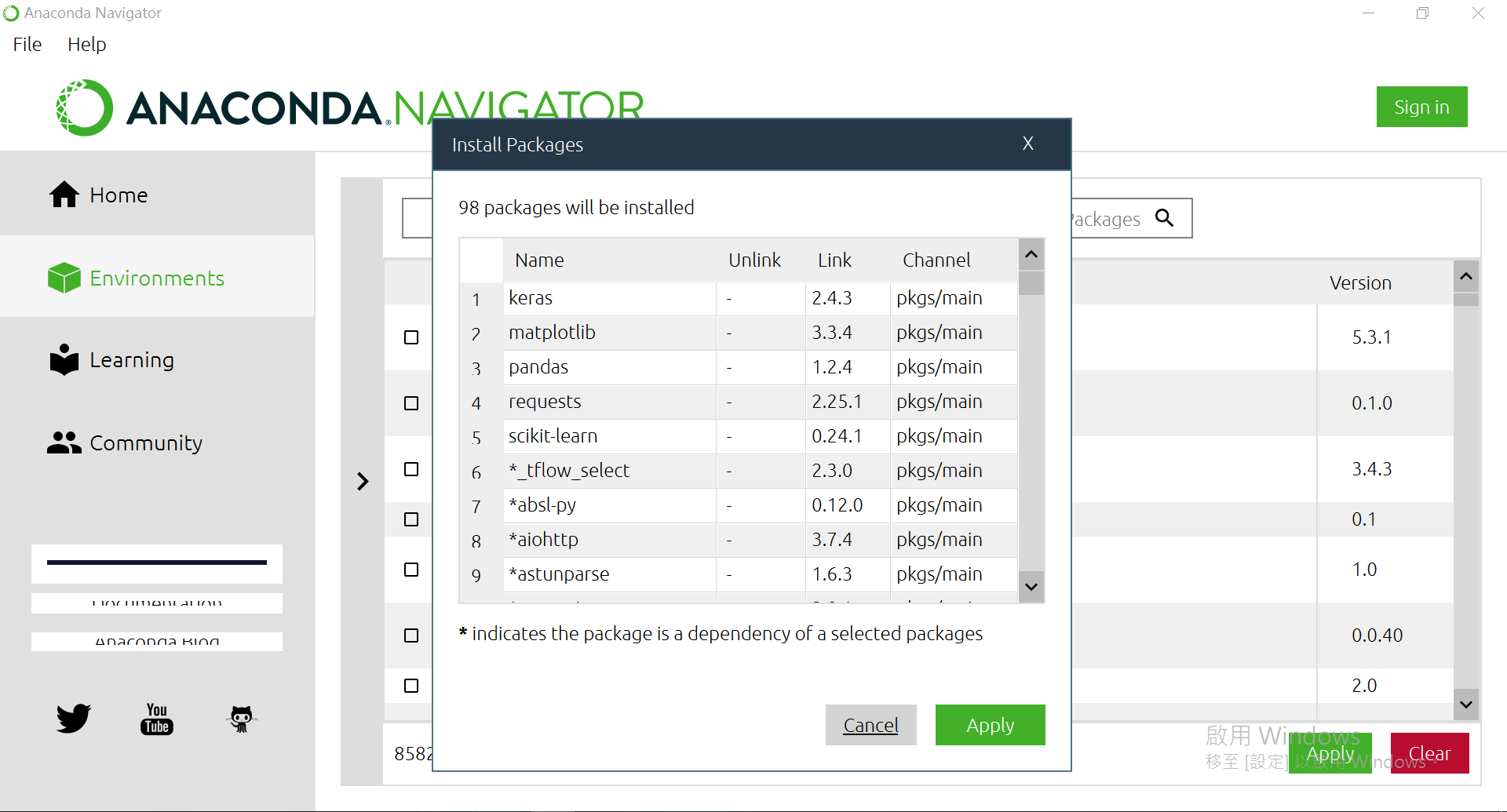
确认过眼神後,再[Apply]一次。
除了透过GUI的套件管理来下载函式库外,也可以用终端机安装。
把箭头按回去後,点一下绿色三角形(记得要确认环境正确喔~不要到时候装错=.=)
点[Open Terminal]来使用pip进行安装。
来安装个套件看看:
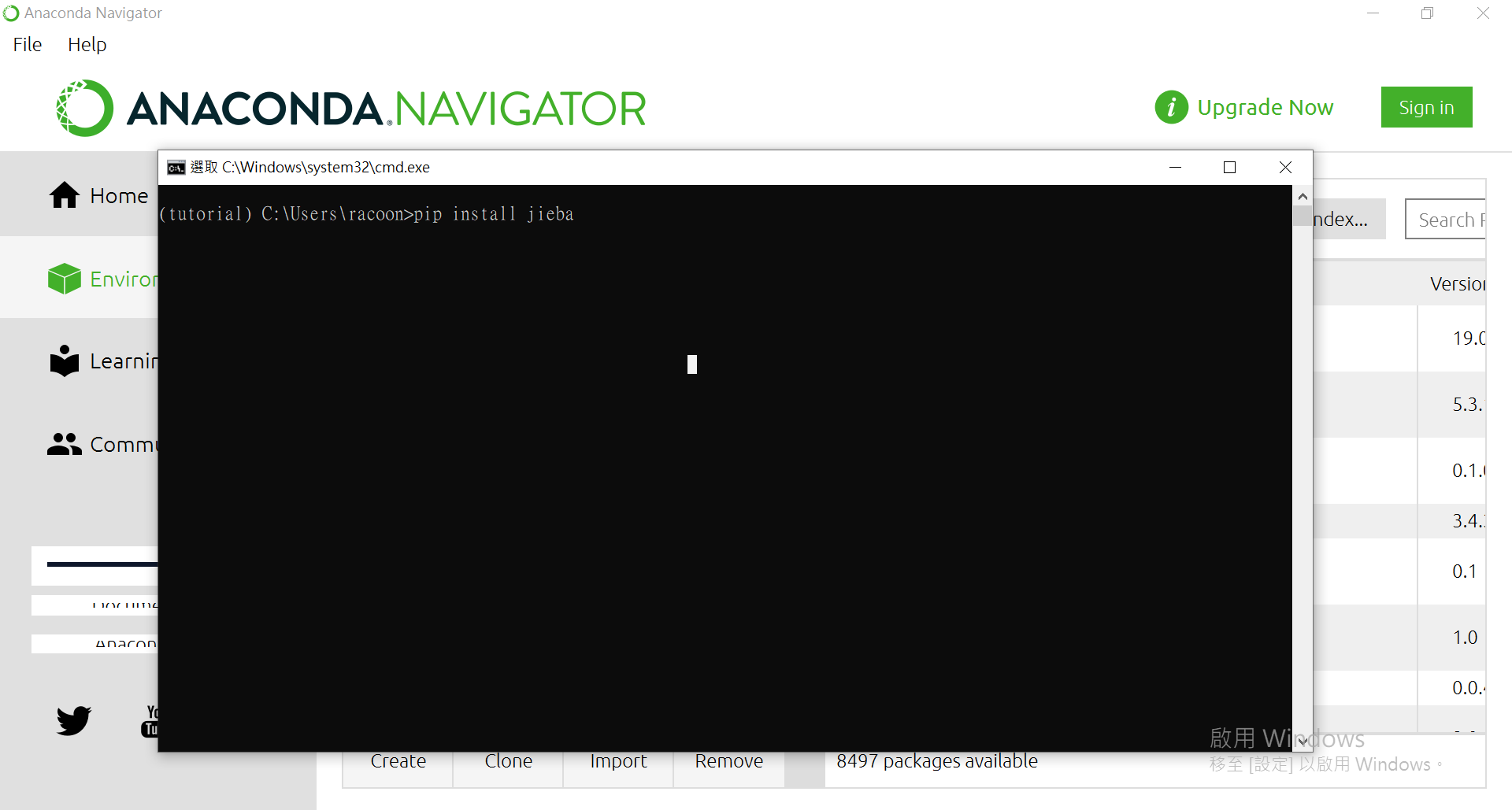
Jupyter Notebook
OK,回到主页。往下滑找到Jupyter Notebook,并按下[Install]
安装完後可以打开。会从浏览器开在本地端的服务器(其实也可以对外远端使用Jupyter)
初始画面:
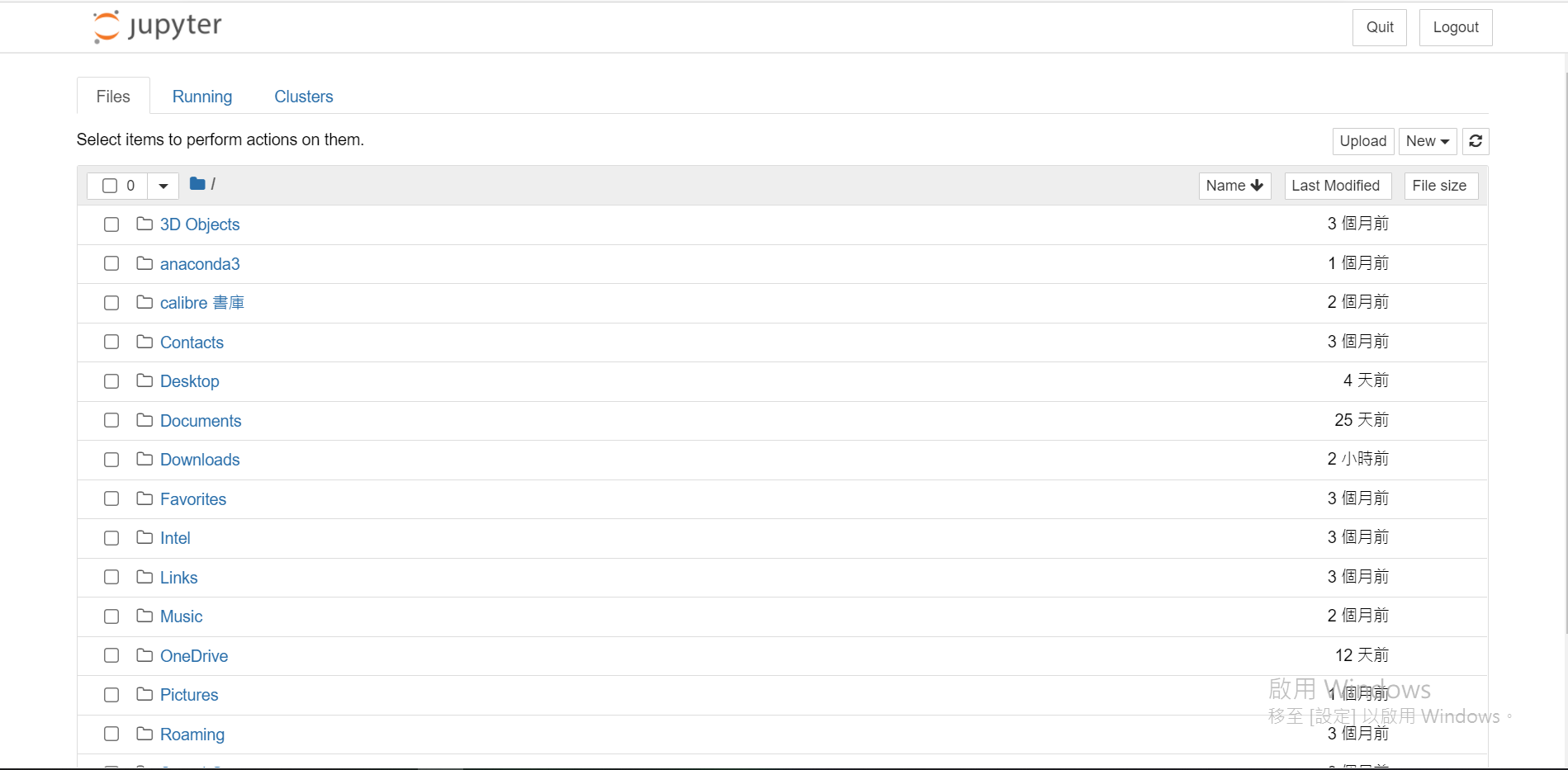
找到/建立一个你喜欢的资料夹(我是放在桌面拉,会叫做Desktop)
进到该资料夹後,右上方有个[New]
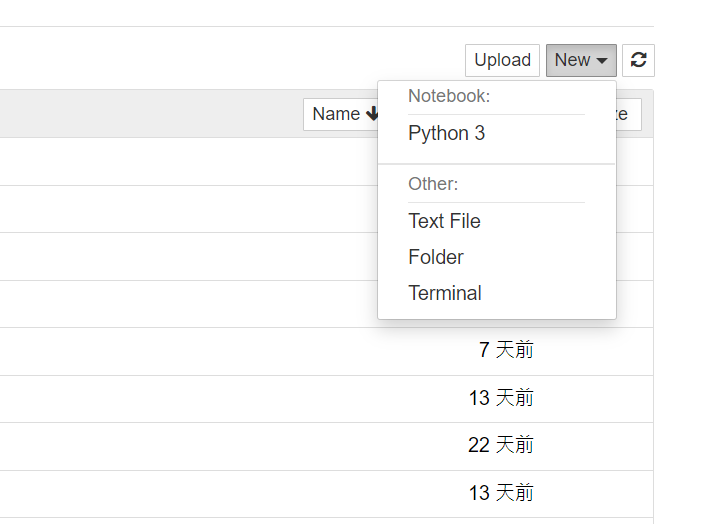
选择Python就可以建立一个ipynb档拉~
机器学习
多元线性回归 Multiple Regression
from sklearn import datasets, linear_model
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
diabetes = datasets.load_diabetes()
diabetes_X = diabetes.data
diabetes_y = diabetes.target
diabetes_X_train, diabetes_X_test, diabetes_y_train, diabetes_y_test = train_test_split(diabetes_X, diabetes_y, test_size=0.2)
model = linear_model.LinearRegression()
model.fit(diabetes_X_train, diabetes_y_train)
diabetes_y_pred = model.predict(diabetes_X_test)
print(f'R2 score: {r2_score(diabetes_y_test, diabetes_y_pred)}')
多项式回归 Polynomial Regression
%matplotlib inline
import matplotlib.pyplot as plt
import numpy as np
from sklearn.preprocessing import PolynomialFeatures
from sklearn.pipeline import make_pipeline
from sklearn.linear_model import LinearRegression
rng = np.random.RandomState(10)
x = 10 * rng.rand(30)
y = np.sin(x) + 0.1 * rng.randn(30)
poly_model = make_pipeline(PolynomialFeatures(7), LinearRegression())
poly_model.fit(x[:, np.newaxis], y)
xfit = np.linspace(0, 10, 100)
yfit = poly_model.predict(xfit[:, np.newaxis])
plt.scatter(x, y)
plt.plot(xfit, yfit)
岭回归 Ridge Regression
import numpy as np
from sklearn import datasets, linear_model
from sklearn.metrics import r2_score
from sklearn.model_selection import train_test_split
diabetes = datasets.load_diabetes()
diabetes_X = diabetes.data
diabetes_y = diabetes.target
diabetes_X_train, diabetes_X_test, diabetes_y_train, diabetes_y_test = train_test_split(diabetes_X, diabetes_y, test_size=0.2)
model = linear_model.Ridge(alpha=1.0)
model.fit(diabetes_X_train, diabetes_y_train)
diabetes_y_pred = model.predict(diabetes_X_test)
print(f'R2 score: {r2_score(diabetes_y_test, diabetes_y_pred)}')
K-NN
from sklearn import neighbors, datasets
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.metrics import accuracy_score
iris = datasets.load_iris()
X = iris.data
y = iris.target
model = neighbors.KNeighborsClassifier()
model.fit(X, y)
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = neighbors.KNeighborsClassifier()
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
决策树 Decision Tree
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.datasets import load_iris
from sklearn.tree import DecisionTreeClassifier
from sklearn import preprocessing
iris = load_iris()
X = iris.data
y = iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = DecisionTreeClassifier()
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
随机森林 Random Forest
from sklearn.ensemble import RandomForestClassifier
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.datasets import load_iris
iris = load_iris()
X = iris.data
y = iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = RandomForestClassifier(max_depth=6, n_estimators=10)
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
支援向量机 SVM
from sklearn.datasets import load_breast_cancer
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
from sklearn.svm import SVC
cancer = load_breast_cancer()
X = cancer.data
y = cancer.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = SVC(kernel='rbf')
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
单纯贝式 Naive Bayes
from sklearn import datasets
from sklearn.naive_bayes import MultinomialNB
from sklearn import preprocessing
from sklearn.model_selection import train_test_split
from sklearn.metrics import accuracy_score
iris = datasets.load_iris()
X = iris.data
y = iris.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2)
scaler = preprocessing.StandardScaler().fit(X_train)
X_train = scaler.transform(X_train)
model = MultinomialNB()
model.fit(X_train, y_train)
X_test = scaler.transform(X_test)
y_pred = model.predict(X_test)
accuracy = accuracy_score(y_test, y_pred)
print('accuracy: {}'.format(accuracy))
深度学习
深度神经网路 DNN
import numpy as np
from keras.models import Sequential
from keras.layers.core import Dense, Activation
from keras.utils import np_utils
#preparing data for Exclusive OR (XOR)
attributes = [
#x1, x2
[0, 0]
, [0, 1]
, [1, 0]
, [1, 1]
]
labels = [
[1, 0]
, [0, 1]
, [0, 1]
, [1, 0]
]
data = np.array(attributes, 'int64')
target = np.array(labels, 'int64')
model = Sequential()
model.add(Dense(units=3 , input_shape=(2,))) #num of features in input layer
model.add(Activation('relu')) #activation function from input layer to 1st hidden layer
model.add(Dense(units=3))
model.add(Activation('relu')) #activation function from 1st hidden layer to 2end hidden layer
model.add(Dense(units=2)) #num of classes in output layer
model.add(Activation('softmax')) #activation function from 2end hidden layer to output layer
model.compile(loss='categorical_crossentropy', optimizer='adam', metrics=['accuracy'])
score = model.fit(data, target, epochs=100)
卷积神经网路 CNN
import numpy as np
from keras.models import Sequential
from keras.layers import Dense, Dropout, Activation, Flatten
from keras.layers import Convolution2D, MaxPooling2D
from keras.utils import np_utils
from keras.datasets import mnist
(X_train, y_train), (X_test, y_test) = mnist.load_data()
X_train = X_train.reshape(X_train.shape[0],28, 28, 1)
X_test = X_test.reshape(X_test.shape[0], 28, 28, 1)
Y_train = np_utils.to_categorical(y_train, 10)
Y_test = np_utils.to_categorical(y_test, 10)
model = Sequential()
model.add(Convolution2D(32, (3, 3), activation='relu', input_shape=(28,28,1)))
model.add(Convolution2D(32, (3, 3), activation='relu'))
model.add(MaxPooling2D(pool_size=(2,2)))
model.add(Dropout(0.25))
model.add(Flatten())
model.add(Dense(128, activation='relu'))
model.add(Dropout(0.5))
model.add(Dense(10, activation='softmax'))
model.compile(loss='categorical_crossentropy',optimizer='adam',metrics=['accuracy'])
model.fit(X_train, Y_train, batch_size=32, epochs=1, verbose=1)
score = model.evaluate(X_test, Y_test)
print('Test accuracy: {}'.format(score[1]))
循环神经网路 RNN
==== 待更 ====
<<: HTTP Token 使用方式: Basic Token v.s Bearer Token
DAY 15 处理接收到的LINE emoji讯息
我这边群组团购讯息会用到很多LINE emoji,这边会将讯息跟LINE emoji做处理 前面Da...
[Day 14]从零开始学习 JS 的连续-30 Days---forEach回圈
阵列 forEach 资料处理方法 语法:宣告阵列的名称+( . )+forEach( + func...
Day6-控制器是在控什麽 controller说明
pod的管理与控制 对k8s来说,pod是k8s最小的元件,但是当我们在使用时,通常不会直接对pod...
【从零开始的 C 语言笔记】第二十三篇-Switch条件式
不怎麽重要的前言 上一篇介绍了两个小题目,稍微带过解题的思路,以及多重回圈(巢状回圈)的概念。 现在...
详解资料仓库的实施步骤,实战全解!(2)
建立资料仓库是一个解决企业资料问题应用的过程,是企业资讯化发展到一定阶段必不可少的一步,也是发展资...